多组差异分析
安装这些包
1 | BiocManager::install(c("openxlsx",'org.Bt.eg.db','GO.db','topGO', |
1 | rm(list = ls()) |
插入表格
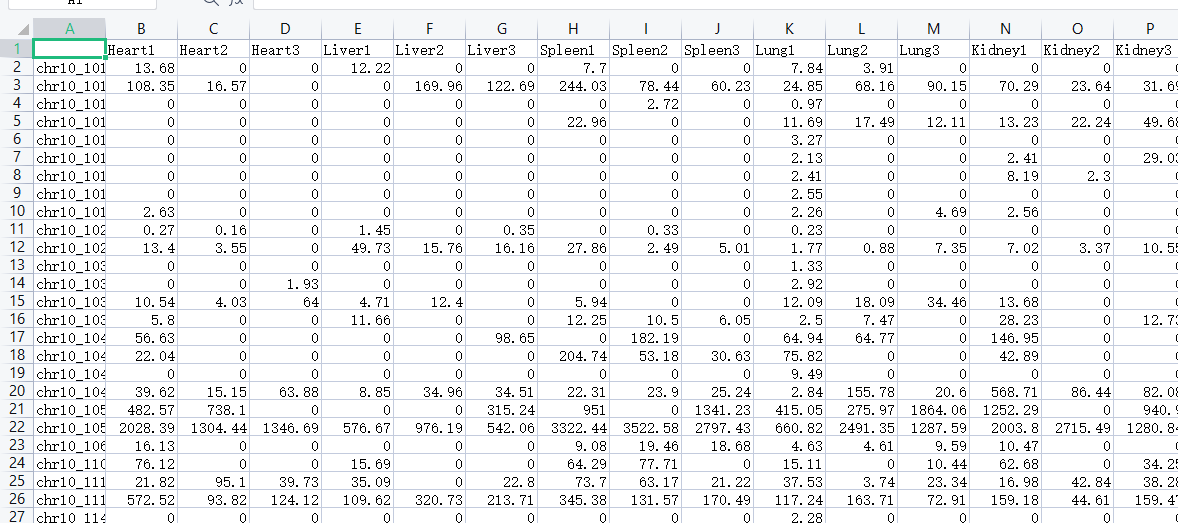
1 | gene_list = read.xlsx("circRNA_ALL_Tpm.xlsx",sheet = 1) |
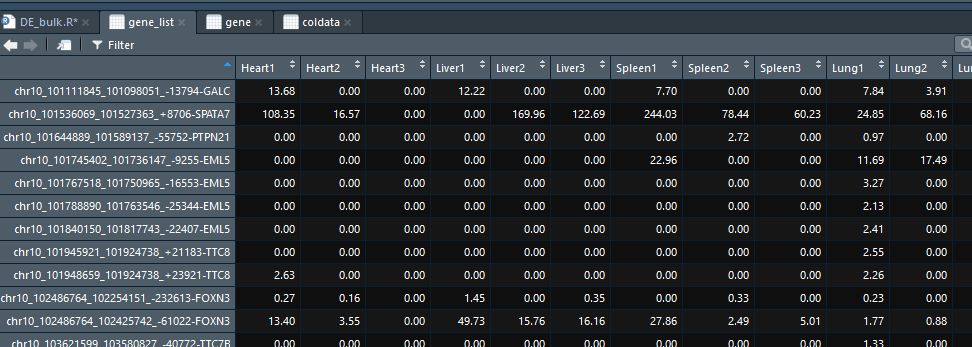
1 | names(gene) |
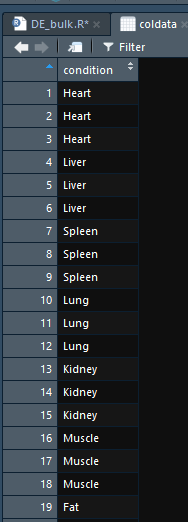
计算
1 | str(coldata) |
循环对比输出!
1 | res_ori <- list() |
本博客所有文章除特别声明外,均采用 CC BY-NC-SA 4.0 许可协议。转载请注明来自 WSZ@blog!
评论
