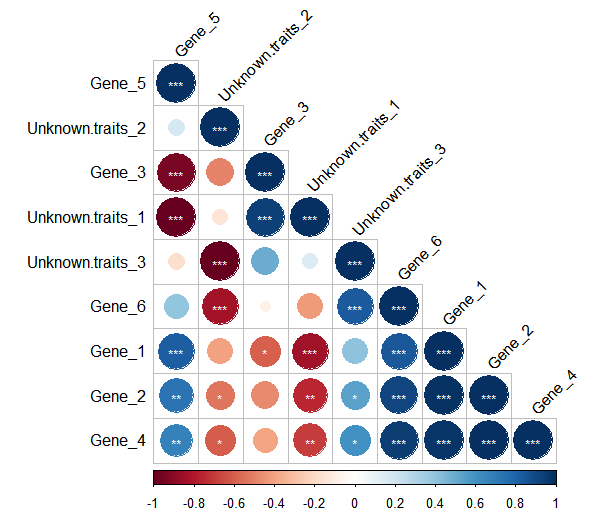
R语言:相关性计算
如果想做成相关性图
不管是基因表达之间的相关性或者是基因与性状之间的关联,可以用R包‘tidyverse’和‘corrplot’实现
首先是录入数据的要求
举个栗子!
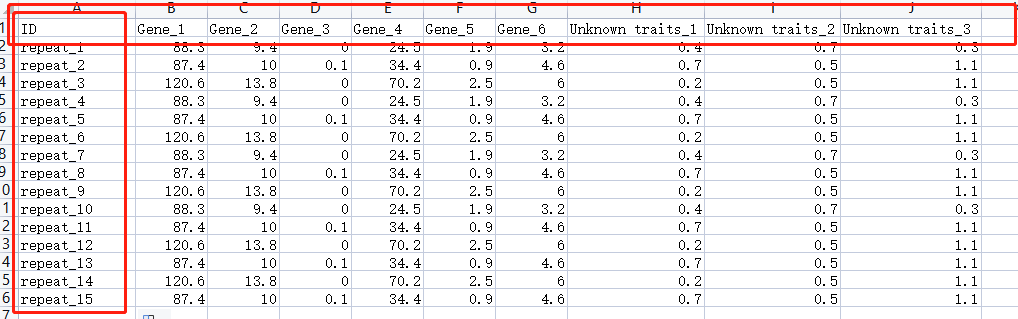
录入数据如上所示
横坐标代表的是所有相关性的基因或者表型或者你需要的其他数据
纵坐标代表重复,重复一般越多越好,三个重复也不是不能做,但是不会出什么好结果,至少重复在4以上
以上数据存储为.csv格式
打开R
代码简单:
不过首先先确定运行文件夹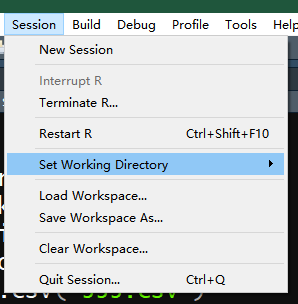
然后导入数据,编辑数据表格
rm(list = ls())
getwd();
workingDir = “.”;
setwd(workingDir);
library(tidyverse)
library(corrplot)
data=read.csv(“example.csv”)
rownames(data)=data$ID
data1=data[,-1]
data=data1
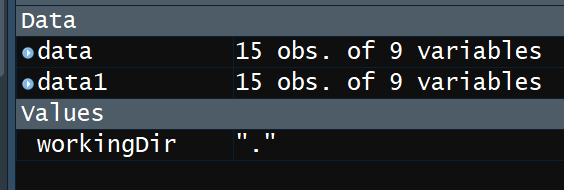
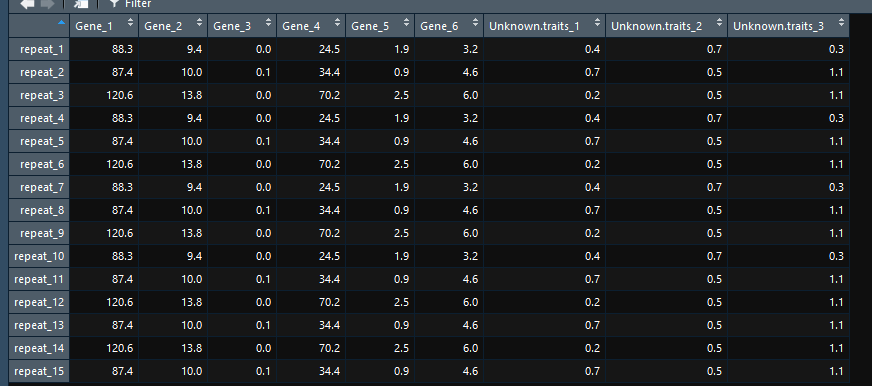
最后data的格式如上!
运行分析代码:
cor(data) %>% corrplot(method = “circle”,order = “hclust”,
type = “lower”,tl.srt = 45,tl.col = “black”)
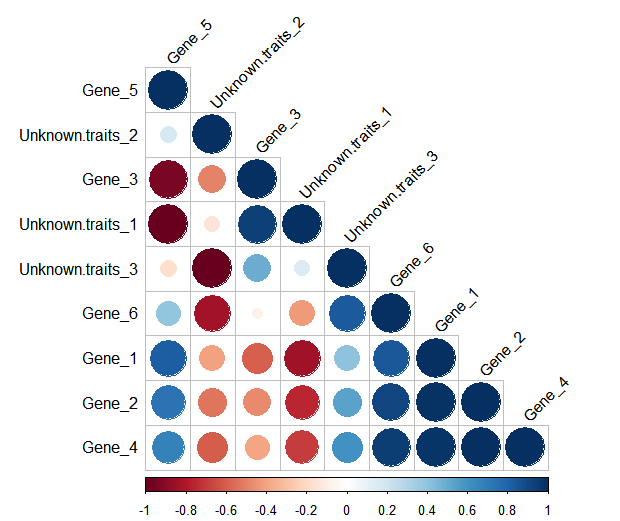
#下面是格式代码如有需要可自行运行
res1 <- cor.mtest(data)
cor(data)%>% corrplot(type=”lower”, order=”hclust”,tl.srt = 45,
tl.col = “black”,
p.mat = res1$p,insig = “p-value”,sig.level = -1)
cor(data)%>% corrplot(type=”lower”, order=”hclust”,
p.mat = res1$p,insig = “p-value”)
cor(data)%>% corrplot(type=”lower”,order=”hclust”,
p.mat = res1$p,insig = “blank”)
cor(data)%>% corrplot(type=”lower”,order=”hclust”,
p.mat = res1$p, sig.level = .05)
cor(data)%>% corrplot(type=”lower”,order=”hclust”,
p.mat = res1$p, sig.level = .01)
cor(data) %>% corrplot(type=”lower”, order=”hclust”,
p.mat = res1$p,insig = “label_sig”,
sig.level = c(.001, .01, .05),
pch.cex = .9, pch.col = “white”,
tl.srt = 45,tl.col = “black”)